Regulation of Pom cluster dynamics in Myxococcus xanthus
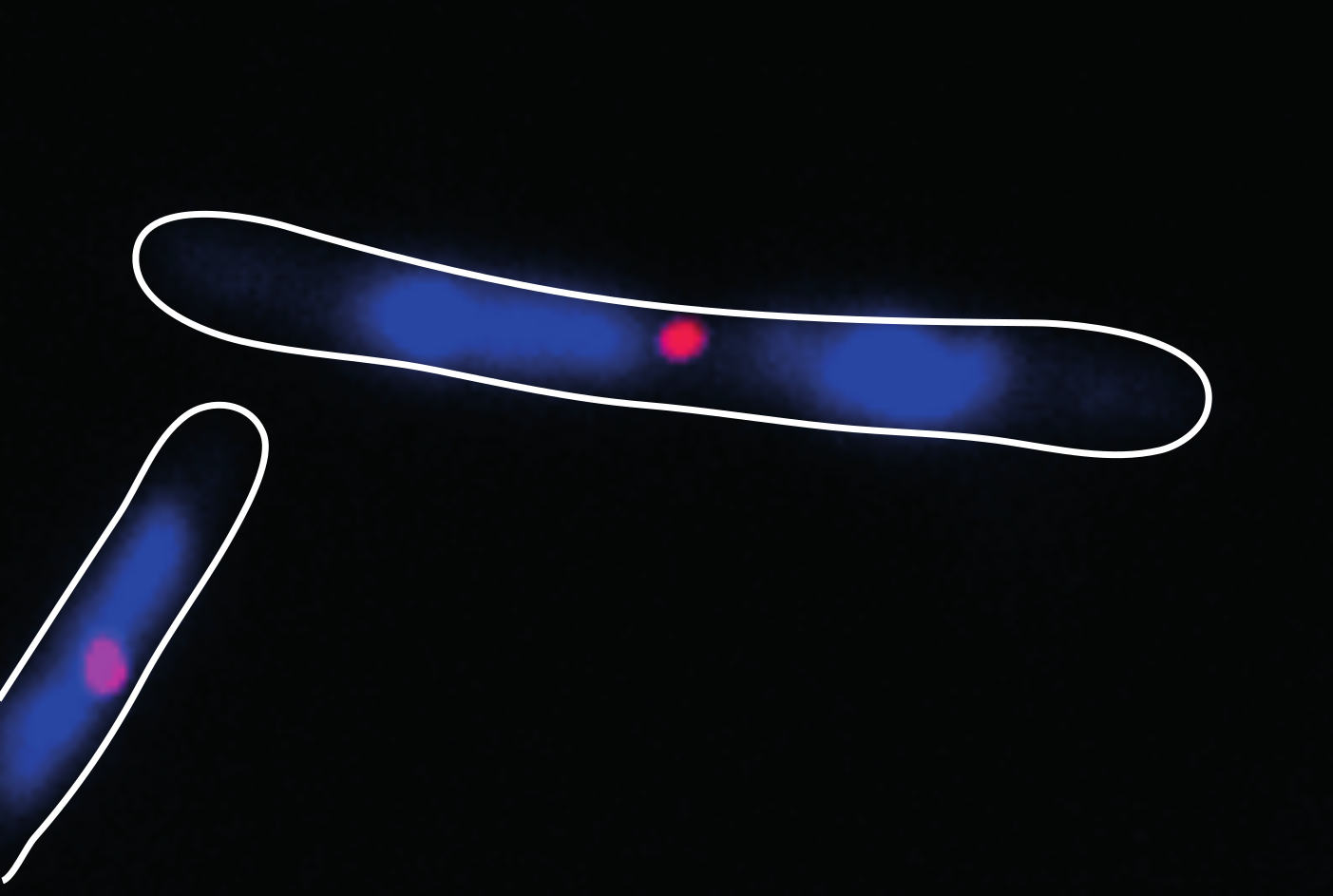 Intracellular positioning of proteins is important for several vital processes in bacterial cells, including cell division. Bacterial cells generally divide symmetrically by forming a contractile ring, which is progressively constricted to form two daughter cells of equal size. An important question to address is how a bacterial cell can identify mid-cell to divide there. In our work we have developed a mathematical model that explains how the plane of division is specified in the rod-shaped bacterium Myxococcus xanthus.
Intracellular positioning of proteins is important for several vital processes in bacterial cells, including cell division. Bacterial cells generally divide symmetrically by forming a contractile ring, which is progressively constricted to form two daughter cells of equal size. An important question to address is how a bacterial cell can identify mid-cell to divide there. In our work we have developed a mathematical model that explains how the plane of division is specified in the rod-shaped bacterium Myxococcus xanthus.
Three proteins have been identified which are required for the proper localization of the plane of cleavage at mid-cell in M. xanthus. Experiments by the research group of Lotte Søgaard-Andersen at the Max-Planck-Institute for Terrestrial Microbiology in Marburg have shown that two of these, named PomX and PomY, assemble to form a large cluster, which will ultimately mark the position of mid-cell. The third, PomZ, is an ATPase – an enzyme that binds the nucleotide ATP and can convert it into ADP. Dimer molecules made of two ATP-bound PomZ proteins can attach to the chromosomal DNA and diffuse along it, and can also bind to the PomXY cluster and diffuse at a lower rate. The action of this system ensures that the cluster is localized to the midpoint of the nucleoid, which coincides with mid-cell, where the contractile ring will form.
In close collaboration with the research group from Marburg we have developed a mathematical model for the Pom cluster dynamics [1]. In the present study [2] we investigated the detailed dynamics that leads to the positioning of the cluster in the center of the nucleoid. The analysis revealed that the PomZ proteins are the crucial components. They first bind to the chromosomal DNA and subsequently recruit the cluster, thus tethering it to the nucleoid. Simultaneous binding of PomZ to the cluster and the chromosomal DNA, however, eventually activates the ATPase activity of PomZ, which causes it to detach from both the cluster and the DNA. It then diffuses in the cytosol and needs to bind ATP and form dimers before it can bind to the nucleoid again. In addition to this delay, one other factor plays an important role in shuttling the cluster to midnucleoid: The chromosome exhibits a certain degree of elasticity, such that a specific position on the chromosome can explore the region around its equilibrium position as a result of thermal fluctuations. Thanks to this elasticity, PomZ proteins that are bound to both the chromosome and the PomXY cluster can exert a net force on the cluster. Moreover, simulations show that the velocity of the cluster depends on the difference between the fluxes of PomZ into the cluster from either side. The crucial point is that, if the cluster is asymmetrically placed, more PomZ proteins will be fed into it from the direction of the longer segment of the nucleoid than from the opposite side. This imbalance in the flux of PomZ serves to push the cluster toward, rather than away from, mid-cell. When the cluster’s location coincides with the center of the chromosome, it remains in place because the number of PomZ molecules impinging on it from each side is essentially the same.
We also studied the effects of the various model parameters on the cluster dynamics in in silico parameter sweeps and made three interesting observations: First, the velocity of the cluster towards mid-cell is optimal for an intermediate rate with which cluster-bound PomZ is released into the cytosol. Second, if the dynamics of PomZ’s movement along the nucleoid is slow relative to the diffusion of the cluster, the latter does not stably maintain its position at midnucleoid. Instead, it oscillates back and forth about the center of the nucleoid. Finally, we found that the more mobile the PomZ dimers are on the cluster consisting of PomX and PomY, the faster the cluster moves towards mid-cell.
Our results are also of interest in the context of other intracellular positioning systems, such as the Min system used to center the contractile ring in E. coli, plasmid segregation, or the mechanisms that are responsible for the localization of flagella. By studying the similarities and differences between the various systems, the general mechanisms on which they are based can be identified.
[2] S. Bergeler, E. Frey, PLoS Comput. Biol. 14(8): e1006358 (2018)
Related Links
- PLoS Comput. Biol. 14(8): e1006358 (2018): Regulation of Pom cluster dynamics in Myxococcus xanthus
- LMU press release (German)
- LMU press release (English)

